- Applications
- DNA Sequencing
DNA Sequencing
Scientists today have a wide variety of methods they can use to explore the how genomic changes or differences impact biology, including whole-genome or targeted sequencing, short or long reads, and numerous methods for assessing chromatin accessibility. By sequencing DNA in a fundamentally new way, the Element AVITI System brings unique strengths to these applications.
Flexible Throughput
The design of the AVITI System offers fast run times and experimental flexibility for DNA sequencing applications. The system has two fully independent, 1 billion read flow cells, giving researchers the choice between per-run throughput of 300 or 600 Gb. Researchers can sequence whole genomes of one or two human trios in 38 hours, or sequence 24 or 48 whole-exome samples in 24 hours with Cloudbreak™ chemistry.
High Accuracy
The AVITI system combines multiple technology innovations to drive notably higher quality data than Illumina systems (Figure 1).

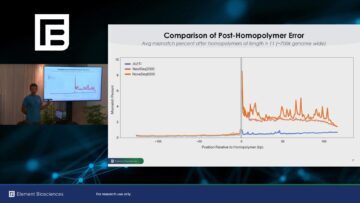
The difference is particularly pronounced in regions following homopolymer regions. As seen in a plot of error rate by genomic position relative to homopolymer start sites, avidite sequencing maintains a consistently low error rate both before and after homopolymer regions (Figure 2A). Another way to look at these data is to compare the ratio of post- versus pre-homopolyer error rates between the two technologies. Evaluating the set of all 700,000 homopolymers in the human genome >11 bp in the vast majority of instances, Element data have a lower ratio of post- versus pre-homopolymer error than Illumina data from the corresponding region (Figure 2B).

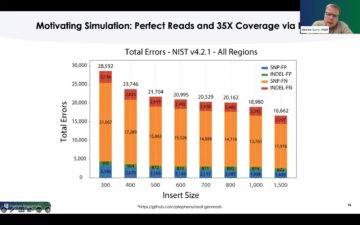
The benefits of higher read quality are seen in fewer variant calling errors in applications such as whole-genome sequencing. A comparison of total SNP and indel errors in 30-fold coverage sequencing of NIST reference genome HG005 demonstrates this advantage (Figure 3). As seen in Figure 3, while both technologies correctly identify heterozygous and homozygous variants in the HG005 truth set, Element does so with greater efficiency, calling fewer false candidates.

The higher efficiency of variant calling carries over to trio sequencing, as shown in a comparison of variants call from the HG002-HG003-HG004 trio on chromosome 20. In trio sequencing, lower coverage of the parent genome can be used to lower the cost of identifying potential causal mutations in cases of undiagnosed disease. In Figure 4, the proband was sequenced to 35-fold coverage while the parents were sequenced to ~17.5-fold coverage (Figure 4). Higher accuracy data limits the number of false do novo variant candidates, even with reduced parental coverage.

High Effective Coverage
Avidite sequencing also leverages technology improvements to produce higher effective coverage that other short-read sequencing platforms, returning more useful data at a given level of coverage. First, the use of rolling circle amplification (RCA) for polony generation reduces optical duplication and eliminates index hopping, limiting the amount of data lost to quality filtering.

Improved imaging instrumentation, including a large field of view with edge-to-edge performance, high resolution optics, and innovative imaging algorithms drive best-in-class performance for low-diversity sequencing. Greatly reduced PhiX spike-in is needed to detect polonies and sustain accuracy across cycles, preserving more throughout for your methylation or amplicon sequencing samples.
Resources