How do I change my Trinity™ hybridization conditions with a custom panel?
Custom panels are enabled with Trinity™, but they may require adjustments to hybridization conditions to maximize performance. Unlike traditional in-solution workflows, Trinity eliminates post-capture amplification, so the molecules captured during hybridization will be directly represented in the sequencing output. As a result, the hybridization conditions provided in the validated workflows for the Twist Bioscience and IDT exome panels do not apply directly to custom panels of different sizes and designs. Contact your Field Applications Scientist or Element Biosciences Technical Support for recommendations when planning Trinity sequencing runs with a custom panel.
While custom DNA probe panels require similar set-up conditions to the validated Twist and IDT exome panels with Trinity, the hybridization input and amount loaded onto the instrument may differ due to the following:
- Panel size and genome size (capture space)
- Probe design
- Probe type
Custom Panels with Trinity: Understanding Hybridization Conditions
One of the most significant factors affecting capture performance with Trinity is the capture space, defined as the ratio of panel size (number of bases covered by the panel without overlap) to the species genome size. Without the post-hybridization PCR that normalizes your number of capture molecules, Trinity requires you to input sufficient targetable library molecules to reach the desired flow cell output. This is done by adjusting the total DNA input into the hybridization reaction, enabling you to achieve the target flow cell output (~800M paired-end reads) with a variety of panel sizes, as shown in Figure 1.
.jpg?width=820&height=485&name=ACT137C_ACT230_Fig1_trinity_panels_alt1_updated%20(1).jpg)
Figure 1 shows the approximate instrument loading amount calculated for a range of theoretical panel capture spaces (grey circles) and tested panel capture spaces (colored circles), which can serve as a helpful starting point for sequencing a custom panel with Trinity. For panels less than 2.5Mb in size, we also recommend end-polishing your Trinity libraries with a few cycles of PCR prior to pooling and hybridization. Pool plexity can be adjusted to meet hybridization input requirements.
Custom Panels with Trinity: Examples
Two example hybridization inputs are shown in Figure 2A for IDT panels of different sizes and designs: the IDT xGen Pan-Cancer panel and IDT xGen Exome v2 panel. To account for large differences in panel size and capture space, the total input into the hybridization reaction and final loading on the instrument is different for these two panels.
.jpg?width=588&height=631&name=ACT137C_ACT230_Fig2_trinity_panels_alt2_updated%20(1).jpg)
|
The smaller Pan-Cancer panel has a panel size of 800kb and a calculated capture space of approximately 0.03% (see example calculation below).
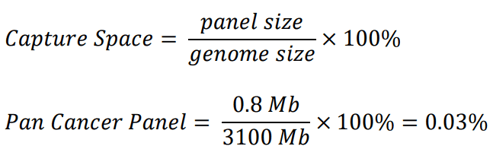
For the smaller Pan-Cancer panel, 24 end-polished libraries were pooled for 24µg total input into the hybridization reaction (1000ng per library). The whole reaction pool was loaded onto the instrument after hybridization and dilution (200µL, or 24µg total instrument loading). This sequencing run yielded 1.07 billion reads, or approximately 44 million reads per sample.
The larger Exome panel has a panel size of approximately 34Mb, and a corresponding capture space of approximately 1.10%. For the larger Exome panel, 24 libraries were pooled for 4µg total input into the hybridization reaction (~167ng per library). About half of this reaction was loaded onto the instrument after dilution (90µL, or ~1.8µg total instrument loading). This sequencing run yielded 1.01 billion reads, or approximately 42 million reads per sample.
Both set-ups achieved similar polony densities of approximately 1 billion reads on a Trinity flow cell and showed comparable or better secondary metrics than their traditional in-solution workflows. As a representative example of this, on-target rate is shown in Figure 2B for each panel. While on-target rate differs with each panel design, Trinity shows a comparable or improved on-target rate for each panel compared to the traditional workflow.
Other factors, such as library size, adapter quality, panel probe design, probe type (dsDNA, ssDNA), and probe amount can also impact output, so some optimization may be required to achieve your target flow cell output for a custom panel. Note that Trinity currently does not support RNA probes, but you can contact your Field Applications Scientist or Support to learn about what other options are available.
Custom Panels with Trinity: Pooling Multiple Hybridization Reactions
All Trinity workflows provide instructions for diluting the hybridization reaction before preparing the final sequencing solution. Once the hybridization reaction is diluted, the correct proportion of the diluted reaction for that panel can be added to the sequencing solution. This amount is determined by your protocol and any customizations you choose to make based on the factors described above.
For some small custom panels, you may have used multiple hybridization reactions to achieve your desired output. In this case, be sure to pool all of the hybridization reactions into the same final sequencing solution, before loading it into the library well. An example of this is provided in the tables below for a custom panel that required two hybridization reactions to be combined:
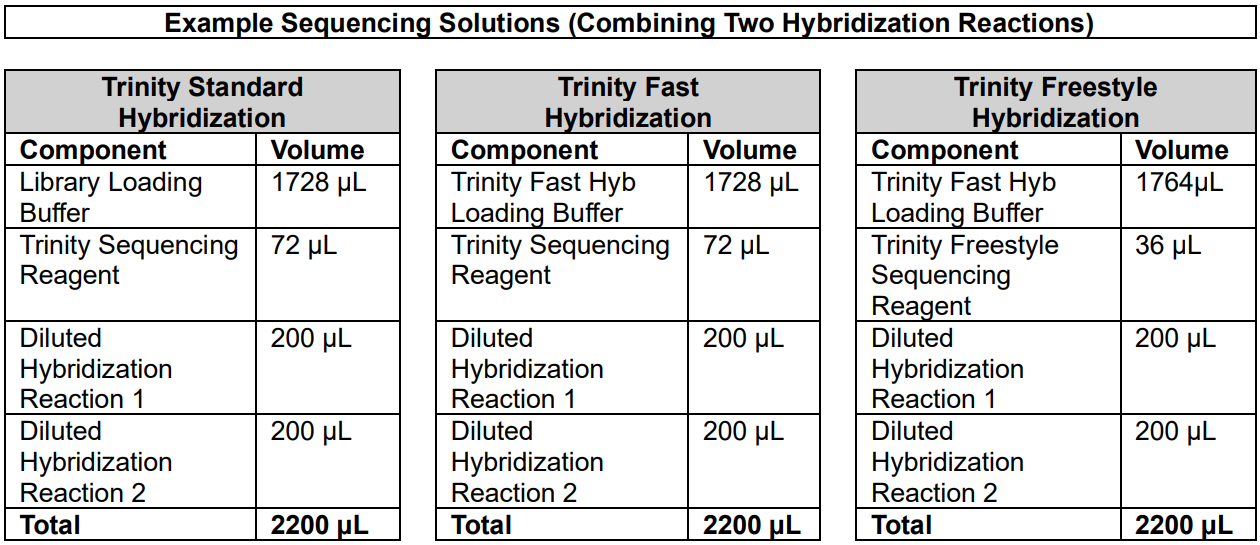
In the above examples, two diluted hybridization reactions are combined in their entirety. To accommodate the additional volume of both reactions, the Loading Buffer amount has been adjusted down. You can further adjust the volume of the Loading Buffer in your workflow to accommodate any volume or combination of diluted hybridization reactions, as long as the final volume of the Sequencing Solution is 2200 µL.
You can learn more about sequencing various panel sizes with Trinity in our pre-print publication, and see an example of sequencing a smaller capture panel in our Application Note: Trinity Workflow for IDT xGen Pan-Cancer Panel.
We recommend contacting your Field Applications Scientist or Support for starting input recommendations prior to setting up your first Trinity sequencing run with a custom panel. Using the hybridization conditions provided in the Trinity exome sequencing protocols as-is with a custom panel is not recommended and may result in lower polony density than expected.
For more information, reach out to your Field Applications Scientist or Element Biosciences Support at support@elembio.com.
Related Articles
We’re here to help — if you can’t find what you’re looking for, let us know.