- Knowledge Base
- Run Setup
- Here
When should I use the Low-Diversity High-Multiplex setting?
This article applies to CloudbreakTM (AdeptTM libraries) or Cloudbreak FreestyleTM (third party libraries)
Introduction
Some applications, like amplicon sequencing, produce libraries with low nucleotide diversity and require unique considerations before sequencing. If the region at the beginning of a sequencing read is highly conserved, it can be difficult to distinguish neighboring polonies that have the same sequence and presents challenges for accurate polony mapping, resulting in lower density than expected. Multiple approaches are available to increase nucleotide diversity at the start of a read, depending on your library and experiment set-up. The Low-Diversity High-Multiplex setting is one approach to increasing nucleotide diversity during polony map generation.
Enabling the Low-Diversity High-Multiplex Setting
Library pools using the low-diversity, high-multiplex setting should meet the following requirements:
- Be third-party libraries sequenced using a Cloudbreak Freestyle sequencing kit or Adept libraries sequenced using a Cloudbreak sequencing kit.
- Have high plexity of ideally ≥ 64 unique sequences in the Index 1 position.
- Use a 1-5% PhiX Control Library spike-in.
- (Note that the Low-Diversity High-Multiplex setting should not be combined with a high PhiX spike-in. This can reduce the index diversity of the pool, leading to a reduction in quality.)
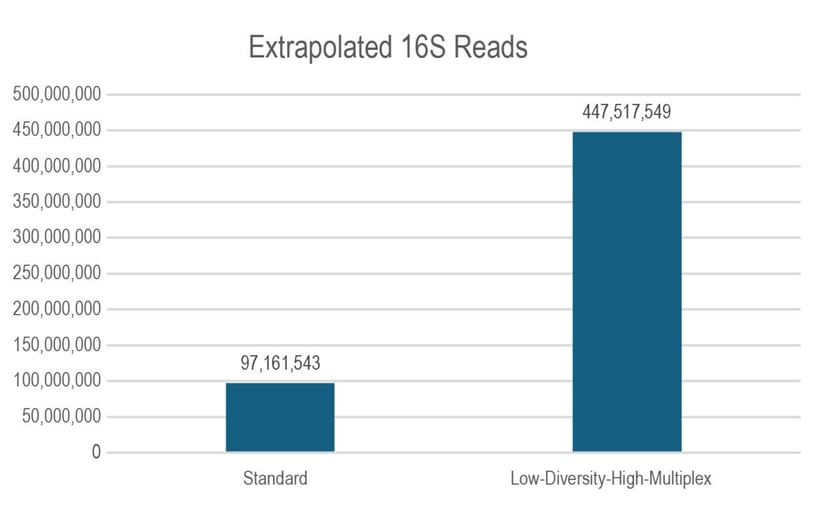
In a test experiment utilizing a 64-plex pool of 16S amplicon libraries, the Low-Diversity High-Multiplex setting more than tripled the total number of 16S polonies detected by the instrument, as shown in Figure 1. For sequencing low diversity libraries without high multiplexing, an alternative approach is required.
Alternative Approaches for Low Diversity Libraries without High Multiplexing
Low diversity libraries that do not meet the criteria for the Low-Diversity High-Multiplex setting require alternative strategies to ensure high quality sequencing. These can include using a higher PhiX control concentration, custom sequencing primers, or phased primers.
High PhiX sequencing strategies use a much higher spike-in of PhiX Control along with normal sequencing settings. This is often combined with a lower library loading concentration, which helps to space out amplicon polonies on the flow cell for better performance. We recommend this strategy for low diversity libraries that do not meet the 64-plex recommendation and thus cannot use the Low-Diversity High-Multiplex setting.
Alternatively, custom sequencing primers or phased indexing primers can be suitable approaches to improve sequencing quality for low diversity libraries, though additional testing and tuning may be required to ensure optimal performance.
For more information, reach out to your Field Applications Scientist or Element Biosciences Support at support@elembio.com.
Related Articles
We’re here to help — if you can’t find what you’re looking for, let us know.