Quantification of Circular Adept™ Libraries Made Simple
Sign up for our newsletter
Join our scientific community to stay up to date with Element news, insights, and product updates.
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.
The Element AVITI™ System uses rolling circle amplification (RCA) for polony generation to decrease error rates caused by other amplification strategies. RCA requires circular library to serve as a template:
- The Element Adept™ Library Compatibility Workflow circularizes libraries on-bench for AVITI sequencing.
- The Element Elevate™ Library Prep Workflow generates a linear library that is automatically circularized onboard the instrument as part of the run.
qPCR quantification versus Qubit
Circular libraries are single-stranded, so the recommendation is to quantify them using qPCR. To support customers who prefer Qubit quantification due to speed and ease of use, Element Biosciences maintains a technical note on how to quantify circular libraries using Qubit.
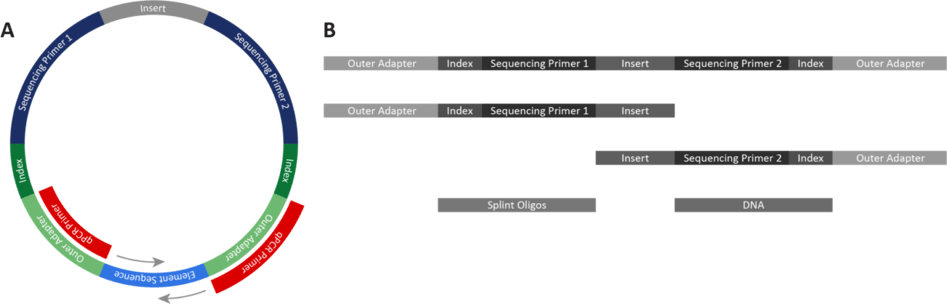
Understanding the qualities of both qPCR and Qubit when determining your routine quantification is essential. qPCR is an accurate method for measuring library concentrations, and is sensitive to lower concentrations. When using the primers supplied in the Adept Library Compatibility Kit v1.1, qPCR detects only circularized library by priming off the outer adapters and targeting the ligation junction of the circularized library during qPCR (Figure 1A).
Using Qubit or other fluorometric quantification methods requires reagents designed for single-stranded DNA (ssDNA). Qubit quantifies any linear DNA contaminating a post-circularization library, including incomplete library fragments, library that did not circularize, primer dimers, splint oligos, and partially digested DNA fragments (Figure 1B). This catch-all quantification can overestimate the library concentration. Furthermore, Qubit cannot detect whether a library has successfully circularized.
Secondary structures in single-stranded circular libraries can overestimate the library concentration, making heat-denaturation necessary before quantification. The technical note documents the procedure. After completing the procedure, you can follow manufacturer instructions to Qubit-quantify the library.
Validating your quantification method of choice
Crucially, validating both quantification methods before routinely quantifying libraries using Qubit verifies that Qubit results are as expected for the library type. A linear relationship exists between libraries quantified using qPCR versus Qubit (Figure 2). Because this relationship is variable, labs might choose to verify Qubit with the libraries they commonly use.
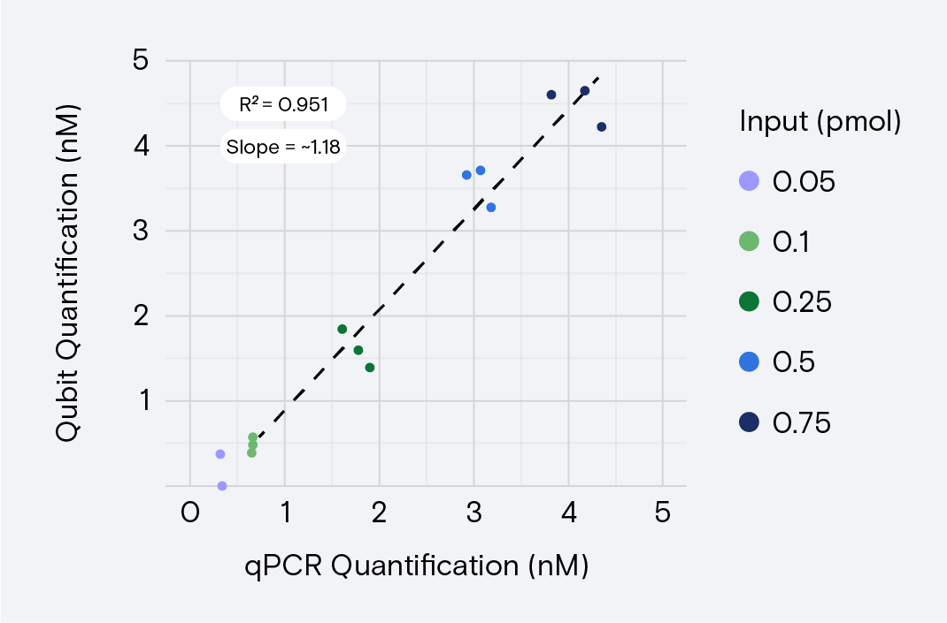
Even after validating Qubit, retaining the ability to quantify with qPCR is important. qPCR results are more accurate than Qubit results and can be used to verify Qubit results, optimize concentrations for new libraries, and troubleshoot unexpected results.
Tracking data about library quality control (QC) and quantification while optimizing Qubit quantifications is helpful. To assist with characterizing the relationship between quantification methods and optimize loading concentration, track the following information:
- Sample type and library prep method
- Linear library QC data, such as quantification results and Bioanalyzer or TapeStation traces
- Adept Workflow input and output concentrations
- qPCR and Qubit quantifications of circularized libraries
- The number of polonies the sequencing run generates
Troubleshooting ligation and digestion issues on Qubit-quantified libraries is accomplished through qPCR, Bioanalyzer, or a fragment analyzer. TapeStation is possible, but it does not always detect incomplete digestion. Figures 3 and 4 provide examples of ligation failure and incomplete digestion for qPCR.
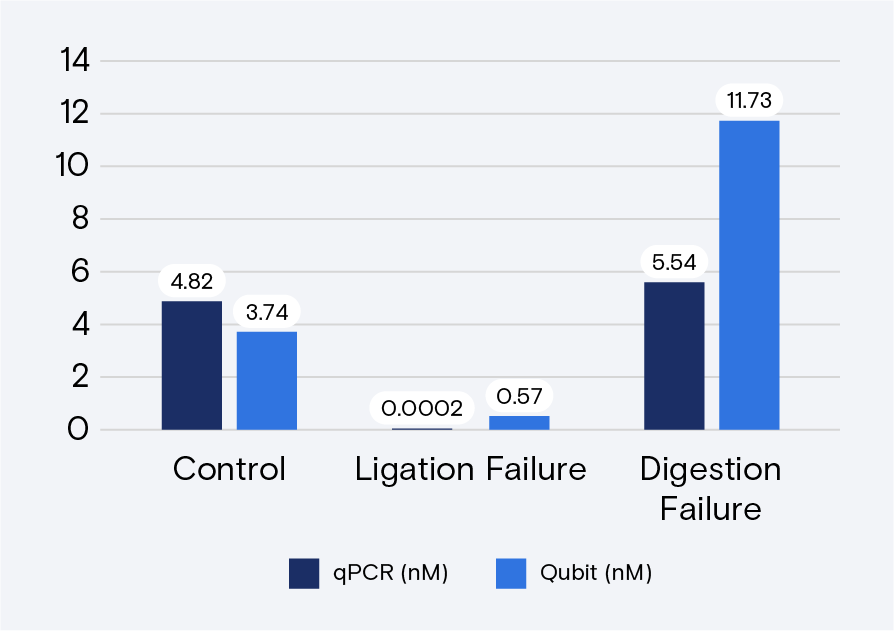
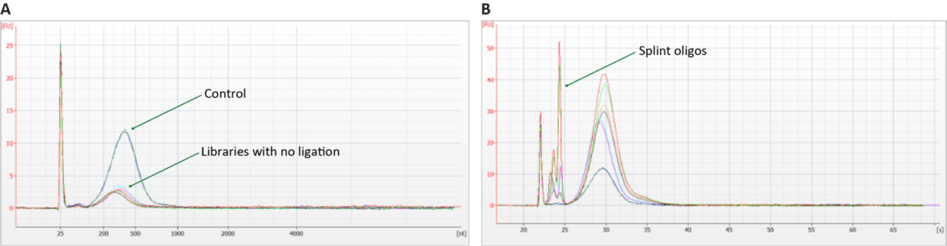
Enabling Qubit quantification for Adept libraries is one of many ways that Element is working to make our industry leading sequencing technology easy to use and accessible to all. To learn more about how the AVITI can help your science now and in the future, contact us here.