Decoding Genetic Variation: De Novo Mutations in Focus
Sign up for our newsletter
Join our scientific community to stay up to date with Element news, insights, and product updates.
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.
How far can precision sequencing take us in understanding the human genome? The answer becomes clearer with each study that pushes the limits of accuracy, scale, and resolution. One of the latest examples is a deep, four-generation analysis, providing a truth set to uncover critical insights into de novo mutations (DNMs) and their implications for genetic variation.
In this Nature publication, authors from the University of Washington School of Medicine leveraged advanced sequencing technologies, including the Element AVITI™ platform, to phase and assemble over 95% of the diploid genomes of the four-generation, 28-member family (CEPH 1463), with near telomere to telomere coverage.

De novo mutations: where variation begins
De novo mutations arise spontaneously, not from inherited DNA, and are a primary source of genetic diversity, a key factor in many rare diseases, and a central focus of rare disease research. Understanding DNMs requires high-resolution phasing, complete diploid assemblies, and the ability to distinguish subtle variation from noise.
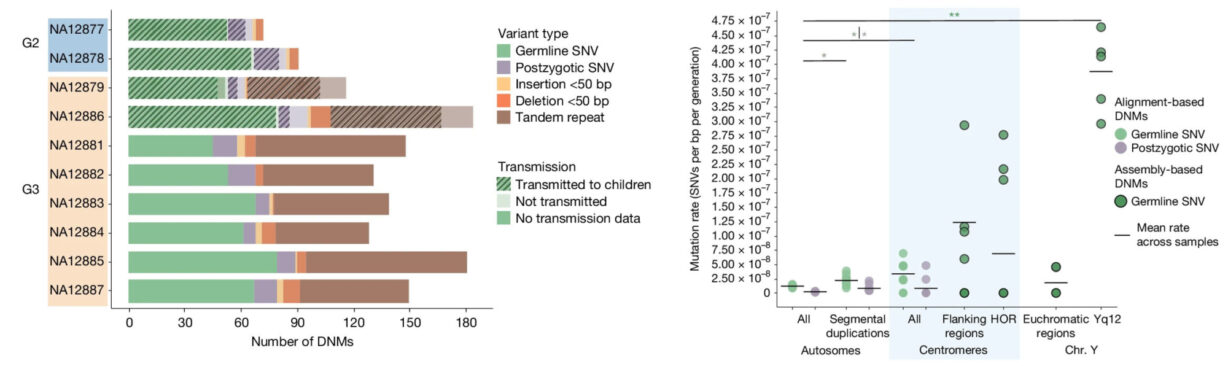
This multiplatform, multigeneration study revealed insights in the repetitive regions, uncovering between 98 and 206 DNMs per transmission, including single-nucleotide variants, non-tandem repeat indels, de novo indels, and centromeric DNMs.
The data revealed a strong paternal de novo bias, demonstrating between 70–80% of germline DNMs originating from fathers and reinforced a potential connection between paternal age and mutation burden.
Interestingly, these mutation types didn’t distribute randomly. Repeats and complex regions that are historically hard to sequence show elevated mutation rates. That’s where high homopolymer accuracy and base-level fidelity really matter.
ABC sequencing through homopolymers
Many DNMs, like de novo tandem repeats, occur in homopolymer or other difficult to sequence regions. The AVITI system leverages our innovative avidite base chemistry (ABC) which uses rolling circle amplification to generate polonies from a single template, creating thousands of copies, so it can provide high-accuracy reads through homopolymers and repetitive regions—areas that are often skipped or miscalled by other technologies.
This study noted the low stutter in Element data at homopolymers and leveraged this high data accuracy to enable precise sequencing of traditionally challenging regions in the genome. By accurately reading homopolymeric areas, the AVITI platform enhanced the detection of mutations and structural variants that were once elusive. This advancement paves the way for deeper exploration of genetic diversity, particularly in regions critical for gene function and disease.
Rethinking inheritance maps
By placing these DNMs on a high-resolution recombination map, the authors tested whether de novo structural variants align with meiotic crossovers. The answer: not always. The expected correlation wasn’t present, suggesting that there is more to learn about the mechanics of recombination and structural change.
The study also found that 16% of de novo SNVs were postzygotic, pointing to early developmental mosaicism. These mutations can complicate interpretation in clinical genetic research and raise new questions around recurrence risk in families.
Looking Forward
The CEPH 1463 study highlights the diversity of human genetic variation and clarifies the mechanisms behind de novo mutations and structural variants. It challenges old assumptions, introduces new questions, and brings the field closer to understanding how genetic variation unfolds.
At Element, we’re proud to see AVITI powering the kinds of studies that make the future of genetics a little clearer. As we advance in our understanding of genetics, we continue to appreciate the complexity of our biology and what it means for health and evolution.