An ability to simultaneously interrogate multiple biomarkers from samples with high heterogeneity—as is the case with tumor samples—has driven rapid uptake of next-generation sequencing (NGS) across the field of oncology research. Therapies that leverage your own immune system and compounds that target specific oncogenes have also shifted the field. The development of novel methods to identify the most effective treatment regimen based on someone’s unique tumor profile are an opportune result. Although research into these companion diagnostics brings great promise to increased adoption of NGS-enabled multigene testing, the cost of sequencing larger comprehensive genomic profiling (CGP) panels hinders adoption and stifles development.
A team at Burning Rock Dx sequenced libraries prepared with their CGP panel, OncoScreen Plus, on the Element AVITI™ System and Illumina NovaSeq 6000 System and compare the results in a preprint. Burning Rock had already validated NovaSeq, so the goal was to determine whether the team benefitted from the flexibility and improved costs of AVITI without impacting results.
Two studies, comparable results
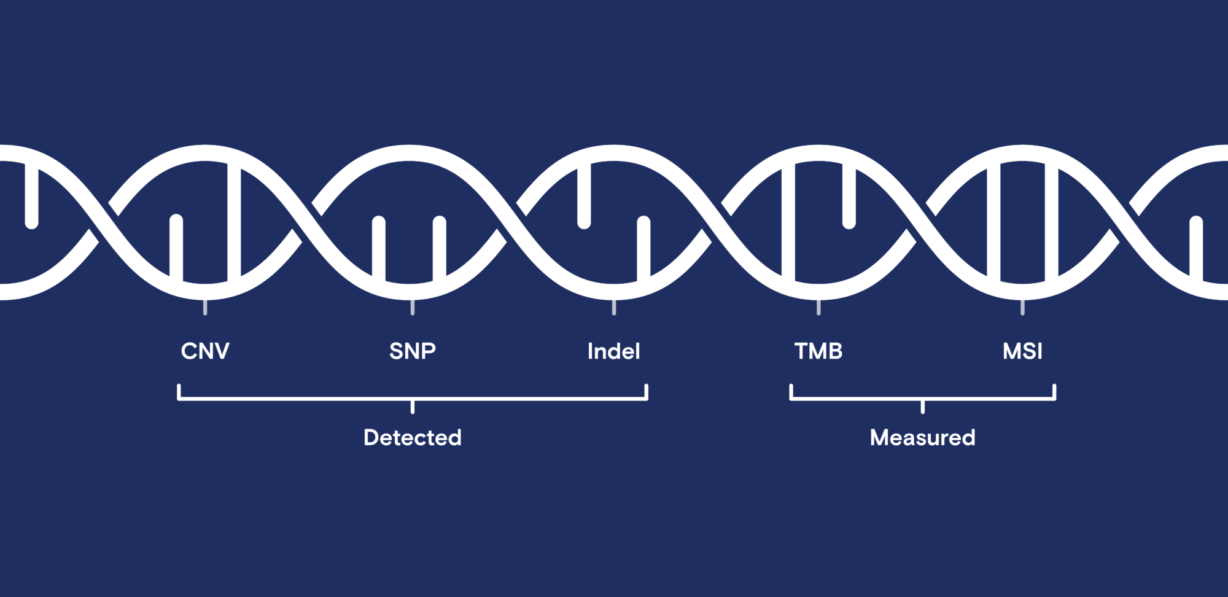
The team performed two research studies to ensure that all reported results were comparable. Each study comprised a comprehensive analysis of known and novel CNVs, SNPs, and indels and complex biomarkers such as microsatellite instability (MSI) and tumor mutation burden (TMB).
The first study leveraged a set of contrived control samples—which allow greater control of large datasets—from a cell line harboring well-characterized variants at known frequencies from ~1% to > 90%. Diluting the sample with a germline control at factors of 1/2, 1/4, 1/8, and 1/16 generated a dataset comprised of hundreds of variant/allele frequency combinations, allowing Burning Rock to determine the concordance levels in detection and relative predicted frequency for both systems. Results show high concordance, detecting the 69 known variants in the control sample and 353 variants detected across the targeted region of the panel.
The second study focused on comparing the results of real-world tumor samples from FFPE blocks. Eight FFPE controls harboring a set of known variants were sequenced on AVITI and NovaSeq. Results indicated nearly identical performance with correlation coefficients > 0.9 for five of the eight samples. Only one sample resulted in 0.75 because of an outlier data point. Additionally, all 35 CNV calls that NovaSeq detected were identified and given the same copy number call as AVITI.
AVITI amplification advantages
In addition to detecting genomic alternations, the OncoScreen Plus assay generates results for complex biomarkers, allowing further comparison across both systems. Unsurprisingly, from both tumor mutation burden and microsatellite instability calculations, all samples in both studies produced highly concordant results.
Remarkably, the authors note a consistently higher average coverage of short tandem repeats (STRs) in AVITI data versus NovaSeq. Similar results observed in Element Biosciences studies are primarily attributed to reduced error propagation during amplification. NovaSeq and most other systems built on sequencing-by-synthesis (SBS) chemistry rely on PCR-based clonal amplification methods such as bridge amplification. AVITI employs rolling circle amplification (RCA) to innovate significant accuracy advantages. RCA continuously copies the original template to improve quality across regions that typically result in PCR stutter, such as homopolymers, STRs, and other repeat elements.
Observation of optical duplicates revealed an additional RCA advantage, with AVITI showcasing fewer optical duplicates. The unbound nature of bridge amplification can lead to the migration of template molecules across regions of the flow cell and colony duplication. The duplicates are later removed from results but can still account for a significant amount of data and reduce usable results. In this case, the Burning Rock team removed ~5–10% of Illumina reads. The tethered Element approach prevents optical duplication and permits almost no data loss.
A trailblazing CGP comparison
The Burning Rock study is the first study to compare CGP results on Element and Illumina systems, and it will not be the last. Given the significant cost savings and flexibility gains to reduce batching constraints, the field is anxious to explore alternatives to current sequencing solutions.
Moreover, the authors note that transitioning their workflows from Illumina to AVITI was virtually seamless and they ran identical libraries on both systems. This assessment includes the analysis pipeline, which was unmodified for analysis of AVITI results. The team expects that further optimization of analysis pipelines for AVITI data can correspondingly push quality even further.